EnviroMail 142 Australia - Detection of Antimicrobial Resistance Genes in Environmental Samples via NGS
For decades, people across the world have relied on antibiotics daily for treating and preventing infections.
Introduction
For decades, people across the world have relied on antibiotics daily for treating and preventing infections. Antibiotics and other antimicrobials have become ingrained in our health and food systems, and our societies. We are now heavily reliant on their effectiveness however, a current rise in antimicrobial resistant and multi-resistant microorganisms is looming as a threat to human and animal health worldwide.
Countermeasures are being developed to reduce the risks posed by this increasing prevalence of antimicrobial resistance (AMR) in the environment. These measures include identification of critical points of control, and implementation of reliable monitoring and surveillance procedures.
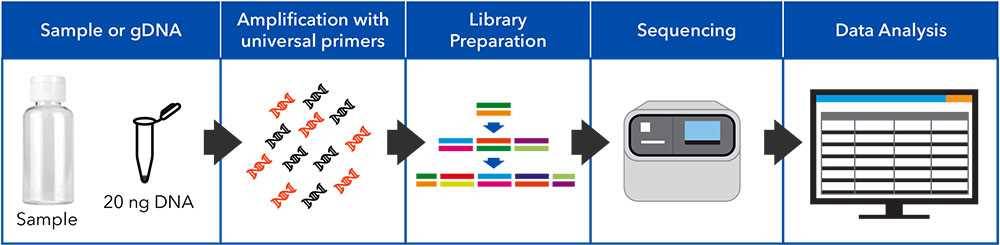
The workflow for detecting AMR genes (ARGs) via Next Generation Sequencing (NGS) is designed to estimate the relative abundance of AMR genes present in a sample by sequencing amplicons from a primer panel targeting more than 500 ARGs.
ALS provides ARG detection in all water matrices (including potable, wastewaters & recycled water) and solid matrices (including sludge, soil, and biosolids).
Guidelines
National strategies to address AMR are still in the early stages of development however many frameworks have been developed globally. The detection of ARGs via NGS plays an important role in each.
Detection of ARGs via NGS provides a valuable and important screening tool that can be applied to a range of applications, particularly for monitoring and surveillance throughout the water, health, agricultural and food industries.
Method and Reporting
Method Code
MM329
Reporting
List and graphical summary of gene families and drug classes detected, by relative abundance (%), based on total read counts.
Principle
An Ion AmpliSeq NGS panel targeting 518 key ARGs was developed in 2018 to investigate the ‘resistome’ on the International Space Station (Urbaniak et al. 2018).
The panel consists of two primer pools that can generate a total of 1,358 amplicons. These primer pools are used in separate multiplex PCRs and the amplicons are prepared for sequencing by ligation of barcoded adapters.
The barcoded libraries are pooled, templated and sequenced using an automated NGS system.
After sequencing, the sequence data are rarefied to a standard read depth, then analyzed with a proprietary ALS analysis pipeline. The pipeline aligns sequences to the ARG reference database and generates a report summarizing the ARG diversity in the sample.
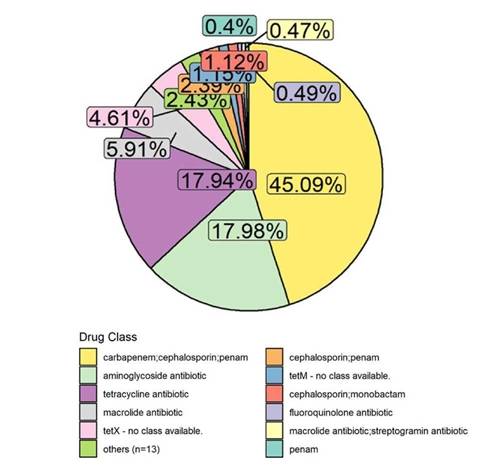
Fig. 1: Pie chart of drug class relative abundances.

Fig. 2: Pie chart of gene family relative abundances.
Industry Applications
Detection of ARGs via NGS can be applied to all water, sewage, soil & sludge matrices. Extracted genomic DNA (gDNA) can be submitted for comprehensive examination of ARG diversity. Industry applications include monitoring and characterization of ARG diversity in:
- Water distribution or treatment processes;
- Urban and agricultural runoff;
- Hospital outflows;
- Agricultural materials and products;
- Water system surveillance for regular monitoring at strategic points.
Detection of Antimicrobial Resistance Genes Analysis
There are three main options for analysis of AMR genes in the environment: NGS-, qPCR-, and culture-derived. Of these, NGS can provide the greatest wealth of information, and is the preferred analysis when total ARGs, ARG diversity, clinically relevant ARGs, mobile ARGs, emerging ARGs, or resistome risk score are being investigated (Liguori et al. 2022).
Thousands of unique ARGs have been identified in bacteria, conferring resistance to a large range of antimicrobial drug classes, such as carbapenems, aminoglycosides, tetracyclines, and macrolides. ARGs can be further grouped into gene families based on how their coded structures confer resistance. A range of mechanisms exist for how ARGs confer resistance, such as efflux pumps that export substrates directly, or enzymes that inactivate the drug.
The NGS option for detecting ARGs can be performed on samples from all types of water and solid matrices. After collection, DNA is extracted using robotic extraction systems. Two separate multiplex PCRs are performed using specific primer pools and the amplicons are prepared for sequencing by ligation of barcoded adapters.
The barcoded libraries are pooled, templated, and sequenced using an automated platform. The sequence reads are quality trimmed and rarefied before being aligned to the ARG reference database, and a report is generated summarizing the ARG diversity in the sample.
Results for each sample include a list of drug classes and gene families observed in the bacterial community, ranked by relative abundance, along with pie charts, see figures, summarizing this information. Raw sequencing files and data are available upon request. QC samples and stringent quality checks along each step of the workflow ensure that output data meets the highest quality standards.
This test approach is sensitive, uses small sample size, and has large sample throughput capabilities. This analysis is a valuable tool for monitoring of ARGs circulating in human populations in wastewater, recycled water, surface water, urban runoff, failed sewage, biosolids, agricultural runoff, and hospital outflows. The insights from this test can help assist in better management and decision making.
Sampling Requirements
| Holding time | 72 hrs for samples stored at 5°C ± 3°C |
| Turnaround time | Standard: 20 business days (5 business days available if required) |
| Sample volume |
Low turbidity water matrix: 2 L in sterile bottles |
| Sample shipping and storage | Transport sample on ice or in a refrigeration unit. |
References
Urbaniak C, Checinska Sielaff A, Frey KG, Allen JE, Singh N, Jaing C, Wheeler K, Venkateswaran K (2018) Detection of antimicrobial resistance genes associated with the International Space Station environmental surfaces. Sci. Rep. 8: 814.
Liguori K, Keenum I, Davis BC, Calarco J, Milligan E, Harwood VJ, Pruden (2022) Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control. Environ. Sci. Technol. 56(13): 9149-9160.
Get in touch with us
If you have any questions relating to detection of Antimicrobial Resistance Genes, please contact:
Brisbane
Sydney
Melbourne
Perth